


Next: 3.2.2 Alternative distance measure
Up: 3.2 Extension of Neural
Previous: 3.2 Extension of Neural
3.2.1 Algorithm
In the extension of Neural Gas to NGPCA, a code-book vector is replaced by a hyper-ellipsoid. An ellipsoid has a center 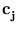 , and axes given by a local PCA, which extracts q eigenvectors
, and axes given by a local PCA, which extracts q eigenvectors
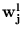 and eigenvalues
and eigenvalues
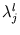 with
l = 1,..., q. The size of the ellipsoid in the direction of the d - q minor components is given by the mean residual variance
with
l = 1,..., q. The size of the ellipsoid in the direction of the d - q minor components is given by the mean residual variance
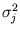 in these directions. The algorithm has the same annealing scheme as Neural Gas, and it also has the same parameters. Again, at the beginning of each annealing step, a data point is randomly drawn from the training set. After presenting a point
in these directions. The algorithm has the same annealing scheme as Neural Gas, and it also has the same parameters. Again, at the beginning of each annealing step, a data point is randomly drawn from the training set. After presenting a point 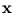 the centers
the centers 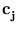 are updated as the code-book vectors in Neural Gas,
are updated as the code-book vectors in Neural Gas,
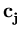 (t + 1) = (t + 1) = 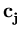 (t) + (t) + 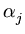 . .  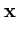 - - 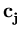 (t) (t)![$\displaystyle \left.\vphantom{{\bf x}- {\bf c}_j(t)}\right]$](img220.gif) . . |
(3.1) |
The weight 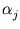 is defined by
is defined by
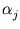 =
= 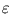 . exp(- rj/
. exp(- rj/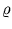 ). The learning rate
). The learning rate
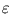 and the neighborhood range
and the neighborhood range 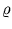 decrease exponentially during training. rj is again the rank of the unit j with respect to
decrease exponentially during training. rj is again the rank of the unit j with respect to 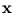 . In the following, the unit index j is omitted for simplicity. All equations need to be evaluated for each unit separately.
. In the following, the unit index j is omitted for simplicity. All equations need to be evaluated for each unit separately.
The ranking of the units cannot depend on the Euclidean distance anymore, since this would ignore the shape of the ellipsoids. Instead, an elliptical error measure is chosen,
E(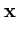 ) = ) = 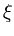 T T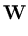 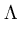 -1 -1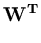 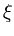 + + 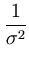 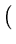 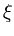 T T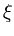 - - 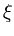 T T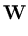 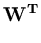 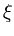  + lndet + lndet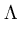 + (d - q)ln + (d - q)ln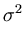 . . |
(3.2) |
This measure is a normalized Mahalanobis distance plus reconstruction error (Hinton et al., 1997).
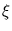 =
= 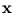 -
- 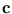 is the deviation of the vector
is the deviation of the vector
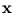 from the center of a unit. The matrix
from the center of a unit. The matrix 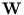 contains in its columns the eigenvectors
contains in its columns the eigenvectors 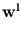 , and the diagonal matrix
, and the diagonal matrix
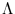 contains the eigenvalues
contains the eigenvalues 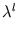 . It can be shown (appendix C.1) that this error measure is the same (up to a constant) as the double negative logarithm of the local probability density given in probabilistic PCA. Thus, a unit can be interpreted as a local density
p(
. It can be shown (appendix C.1) that this error measure is the same (up to a constant) as the double negative logarithm of the local probability density given in probabilistic PCA. Thus, a unit can be interpreted as a local density
p(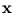 ) = (2
) = (2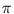 )-d/2exp(- E(
)-d/2exp(- E(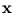 )/2), and the units are ranked in the order of the probabilities of
)/2), and the units are ranked in the order of the probabilities of 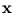 originating from the single units. However, the computation of the probability is not necessary since the same result is obtained by ordering the units using the error
E(
originating from the single units. However, the computation of the probability is not necessary since the same result is obtained by ordering the units using the error
E(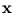 ). The unit resulting in the smallest
E(
). The unit resulting in the smallest
E(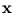 ) has rank zero, and the one with the largest
E(
) has rank zero, and the one with the largest
E(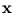 ) has rank m - 1.
) has rank m - 1.
The second term in (3.2) is the reconstruction error divided by 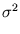 .
. 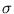 depends on the estimate of the total residual variance
vres, which is updated according to
depends on the estimate of the total residual variance
vres, which is updated according to
vres(t + 1) = vres(t) + 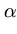 . . 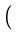 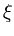 T T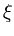 - - 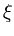 T T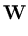 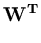 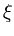 - vres(t) - vres(t)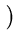 . . |
(3.3) |
The total residual variance is evenly
distributed among all d - q minor dimensions by
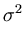 = = 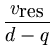 . . |
(3.4) |
This equation is the same as for the noise in probabilistic PCA (2.11).
To adjust the principal axes and their lengths, we do one step with an on-line
PCA method:
{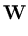 (t + 1), (t + 1),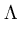 (t + 1)} = PCA{ (t + 1)} = PCA{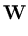 (t), (t),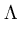 (t), (t),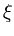 (t), (t),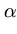 (t)} . (t)} . |
(3.5) |
We use a PCA algorithm similar to the robust recursive least square algorithm (RRLSA) (Ouyang et al., 2000). RRLSA is
a sequential network of single-neuron principal component analyzers based on
deflation of the input vector (Sanger, 1989).
While the 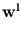 are normalized to unit length, internally the algorithm works with unnormalized
are normalized to unit length, internally the algorithm works with unnormalized
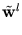 , which are updated according to
, which are updated according to
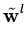 (t + 1) = (t + 1) = 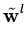 (t) + (t) + 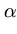 . . 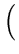 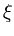 (l)yl - (l)yl - 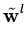 (t) (t)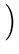 , for l = 1,..., q . , for l = 1,..., q . |
(3.6) |
yl is a component of the vector
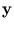 =
= 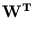
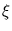 . The deflated vector
. The deflated vector
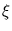 (l) is computed by iterating
(l) is computed by iterating
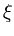 (l+1) = (l+1) = 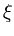 (l) - (l) - 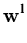 yl starting with yl starting with 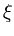 (1) = (1) = 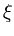 . . |
(3.7) |
After each step t, the eigenvalue and eigenvector estimates are obtained from
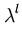 = | = |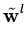 |, |, 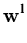 = = 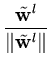 , for l = 1,..., q . , for l = 1,..., q . |
(3.8) |
Since the orthogonality of 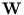 is not fully preserved for each step, the algorithm has to be combined with an orthogonalization method, here we used
Gram-Schmidt (Möller, 2002). Orthogonality is essential for the computation
of the error (3.2). This orthogonalization concludes one annealing step.
is not fully preserved for each step, the algorithm has to be combined with an orthogonalization method, here we used
Gram-Schmidt (Möller, 2002). Orthogonality is essential for the computation
of the error (3.2). This orthogonalization concludes one annealing step.
The unit centers are initialized by
randomly chosen examples from the training set.
The eigenvector estimates are initialized with random orthogonal
vectors. The eigenvalues
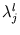 and the variances
and the variances
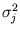 are
initially set to one. To avoid zero variance in a direction during the computation of the local PCA, a uniform noise randomly chosen from the interval
[- 0.0005;0.0005] is added to each component of a randomly drawn data point (for all experiments).
are
initially set to one. To avoid zero variance in a direction during the computation of the local PCA, a uniform noise randomly chosen from the interval
[- 0.0005;0.0005] is added to each component of a randomly drawn data point (for all experiments).



Next: 3.2.2 Alternative distance measure
Up: 3.2 Extension of Neural
Previous: 3.2 Extension of Neural
Heiko Hoffmann
2005-03-22
![]() , and axes given by a local PCA, which extracts q eigenvectors
, and axes given by a local PCA, which extracts q eigenvectors
![]() and eigenvalues
and eigenvalues
![]() with
l = 1,..., q. The size of the ellipsoid in the direction of the d - q minor components is given by the mean residual variance
with
l = 1,..., q. The size of the ellipsoid in the direction of the d - q minor components is given by the mean residual variance
![]() in these directions. The algorithm has the same annealing scheme as Neural Gas, and it also has the same parameters. Again, at the beginning of each annealing step, a data point is randomly drawn from the training set. After presenting a point
in these directions. The algorithm has the same annealing scheme as Neural Gas, and it also has the same parameters. Again, at the beginning of each annealing step, a data point is randomly drawn from the training set. After presenting a point ![]() the centers
the centers ![]() are updated as the code-book vectors in Neural Gas,
are updated as the code-book vectors in Neural Gas,
![]() .
. ![]() depends on the estimate of the total residual variance
vres, which is updated according to
depends on the estimate of the total residual variance
vres, which is updated according to
![]() and the variances
and the variances
![]() are
initially set to one. To avoid zero variance in a direction during the computation of the local PCA, a uniform noise randomly chosen from the interval
[- 0.0005;0.0005] is added to each component of a randomly drawn data point (for all experiments).
are
initially set to one. To avoid zero variance in a direction during the computation of the local PCA, a uniform noise randomly chosen from the interval
[- 0.0005;0.0005] is added to each component of a randomly drawn data point (for all experiments).